RVdb Help Document
Data browsing and retrieval
1.How to Retrieve and Browse RV Strain Data?
2.How to Retrieve and Browse RV Nucleotide Sequence and Protein Sequence Data?
3.How to Retrieve and Browse RV 3D Protein Structure Data?
4.How to Retrieve and Browse RV Related Publications Data?
5.RVdb data retrieval error
Online analysis function
1.How to perform RVdb online multiple sequence alignment?
2.How to build a phylogenetic tree fully automatically on RVdb?
3.How to perform RVdb BLAST analysis?
4.How to perform RV sequence annotation analysis?
5.How to perform rhinovirus molecular typing analysis?
Data browsing and retrieval
1.How to Retrieve and Browse RV Strain Data?
Strain data is of paramount importance in RVdb as it serves as the foundation for generating both nucleotide and protein sequences of rhinoviruses. This data type encompasses comprehensive virus meta information, making it crucial to learn efficient retrieval and browsing methods for virus strain data.
(1) To begin querying Strain data, click on "Data" in the top navigation bar or "Strain" on the home page (the same procedure applies to the other four data types).
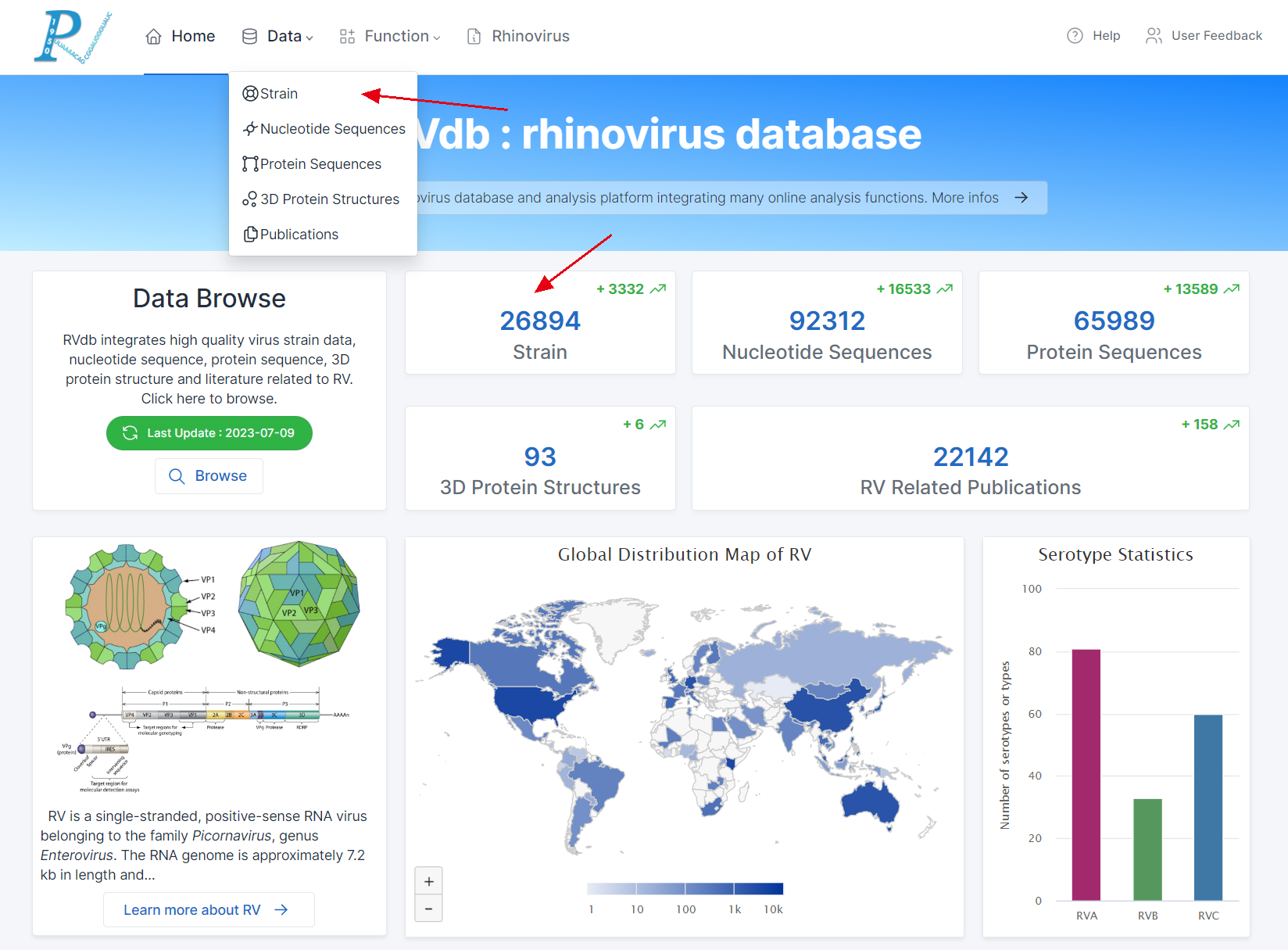
(2) The search form can be freely switched between the five data types.
(3) Users have two distinct search strategies: directed search and screening search.
(4) Directed search: If a user intends to search for a specific rhinovirus strain using the Strain ID (RVdb's unique identifier for virus strains) or GenBank Accession, simply enter the correct Strain ID or Accession in the corresponding input box and click "submit".
(5) Screening search: If a user's objective is not limited to a particular virus strain and instead wants to apply different screening conditions, various parameters can be configured. Note: When input is provided in the directed search input box, it takes precedence, and the form defaults to a directed search submission.
(6) The following example illustrates a search: Retrieving RVA1 strains with VP1 gene collected in the United States since 2020, with a sequence length ranging from 500nt to 7300nt.

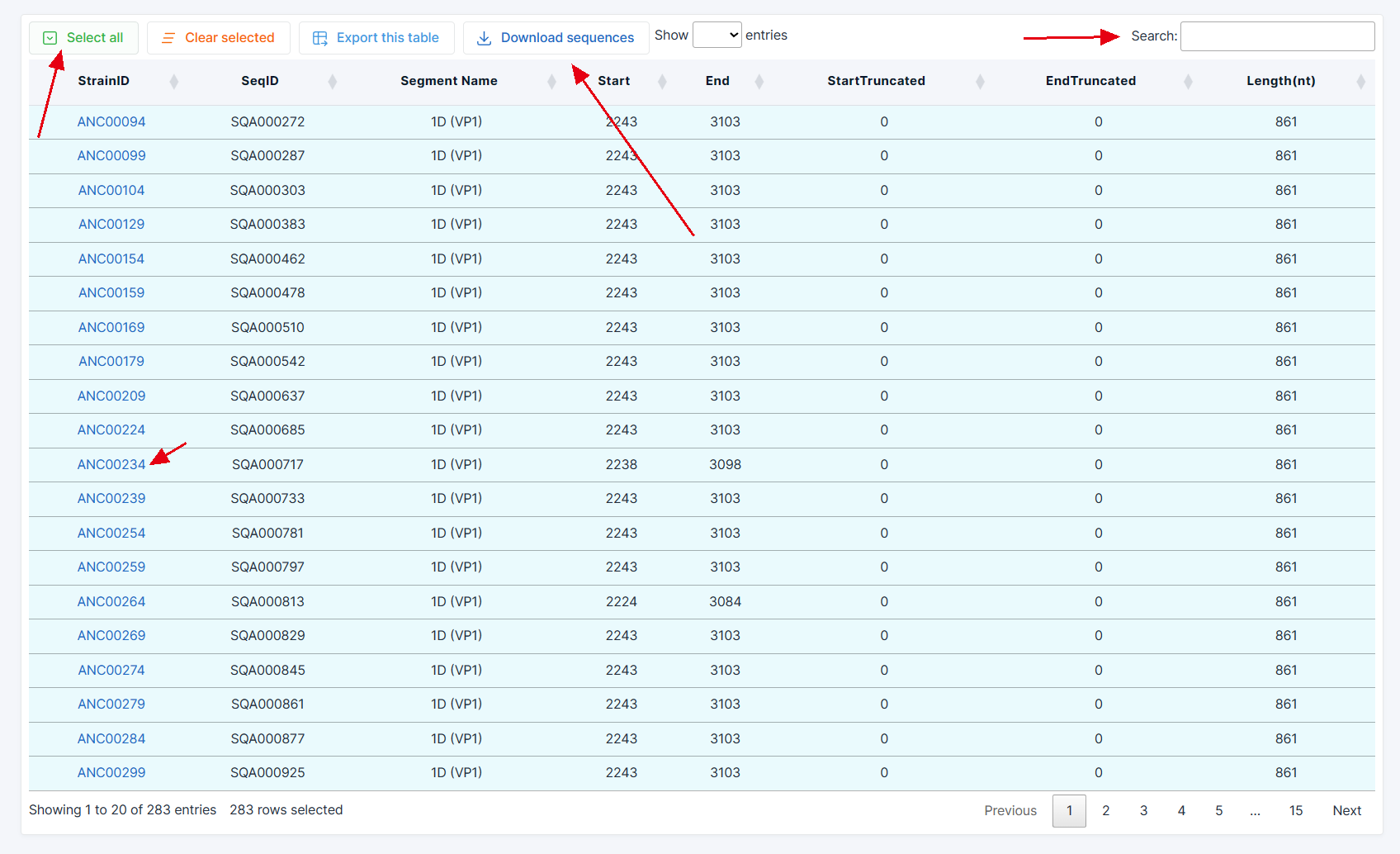
(7) In the result table, users can sort information in each column, search for keywords to query, or download rhinovirus strain data.
(7) In the result table, users can sort information in each column, search for keywords to query, or download rhinovirus strain data.
(8) Additionally, clicking on the Strain ID in the first column allows users to access the virus strain information page for more detailed information on the selected strain.
2.How to Retrieve and Browse RV Nucleotide Sequence and Protein Sequence Data?
(1) The retrieval logic for nucleic acid sequences and protein sequences is identical to that of Strain data, encompassing both directed search and screening search.
(2) Each nucleic acid sequence and protein sequence in RVdb is assigned a unique identifier called Sequence ID. Users can input Sequence ID, Strain ID, or Accession for directed search.
(3) Alternatively, users can perform screening search by configuring other parameters like Strain data searching.
(4) As depicted in the illustration, a search example involved retrieving nucleic acid sequences of the VP1 gene with a length between 300nt and 1000nt from RVA1 strains collected in the United States since 2020.

(5) Within the result table, users have the ability to sort information in each column, search for keywords, or download rhinovirus sequences.
3.How to Retrieve and Browse RV 3D Protein Structure Data?
(1) Currently, there are fewer than 100 available 3D protein structure data for rhinoviruses. Therefore, to ensure a better user experience and more efficient data retrieval, RVdb avoids complex forms for data filtering. Instead, it offers two options: "Browse all RV 3D protein structures" and PDB ID search.
(2) Users can either choose to browse through all 3D protein structures or enter a specific PDB ID to access the corresponding structure.
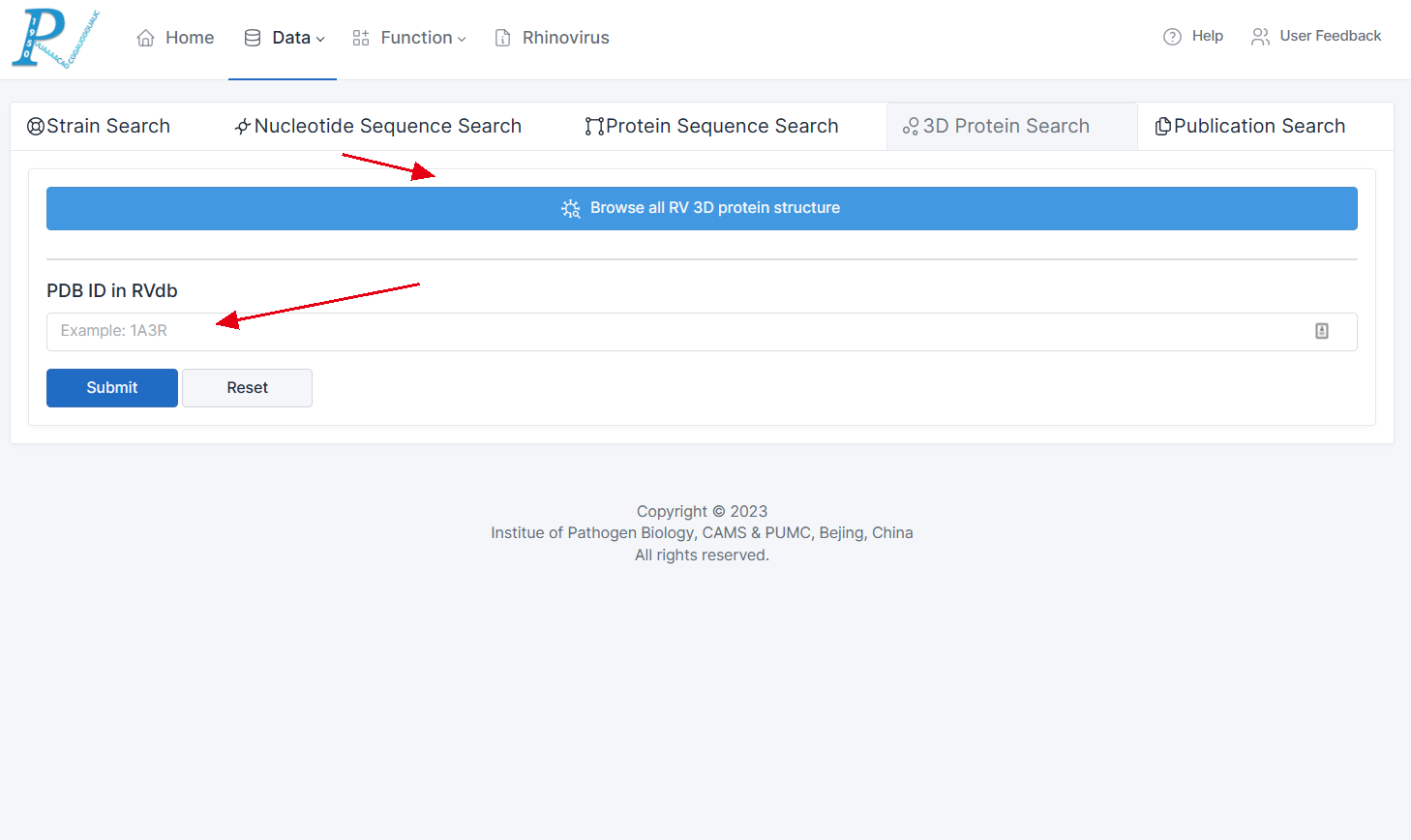
(3) In the result table, users can click on the PDB ID in the first column to access the detailed information page for the specific protein structure.
4.How to Retrieve and Browse RV Related Publications Data?
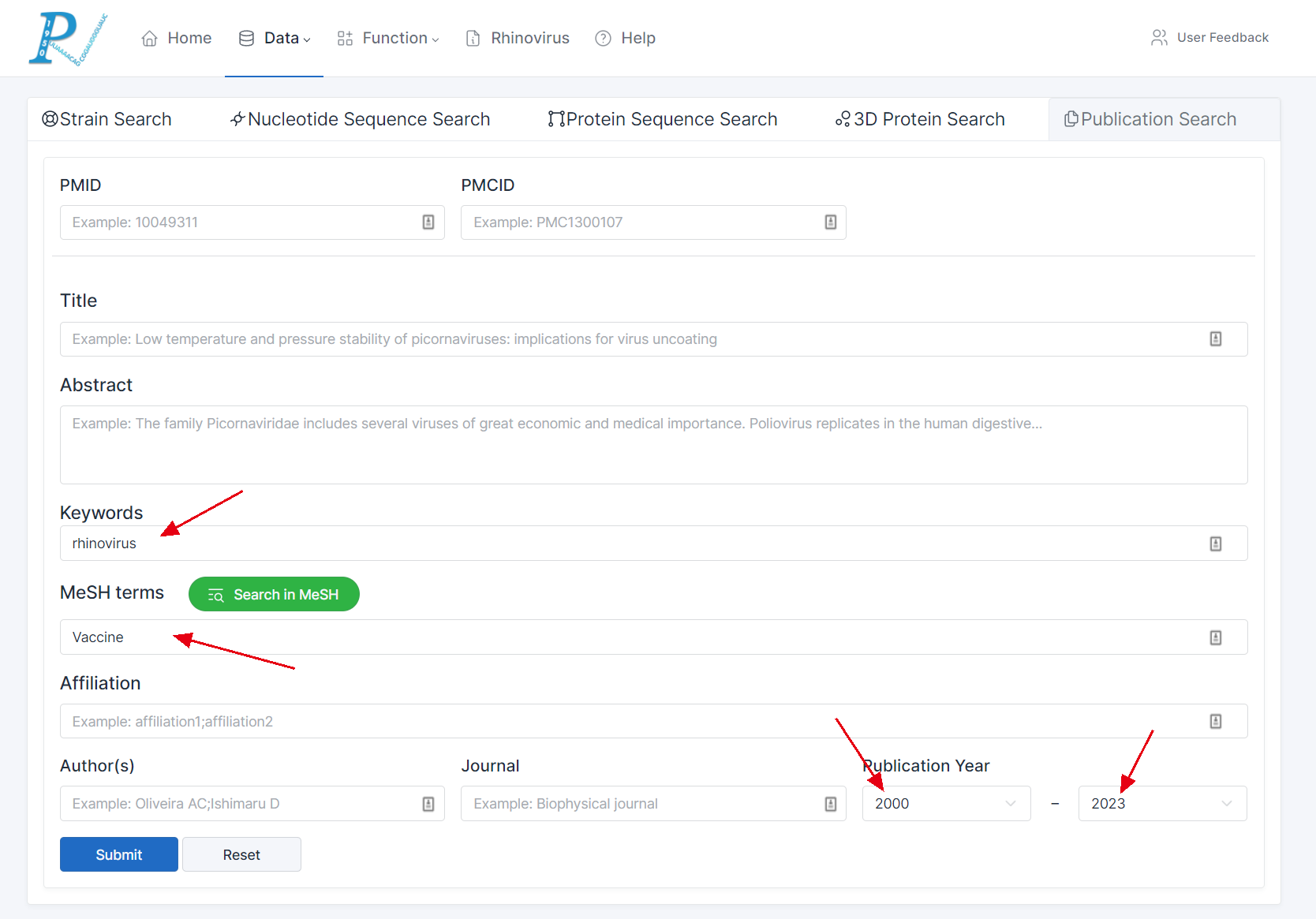
(1) The publication search feature is also divided into direct search and screening search methods.
(2) Direct search enables users to input a specific PMID or PMCID to target a particular document.
(3) Users can perform searches by entering information or parameters such as "Title," "Abstract," "Keywords," or "MeSH terms".
(4) The returned result table provides sorting, filtering, and keyword filtering functionalities.
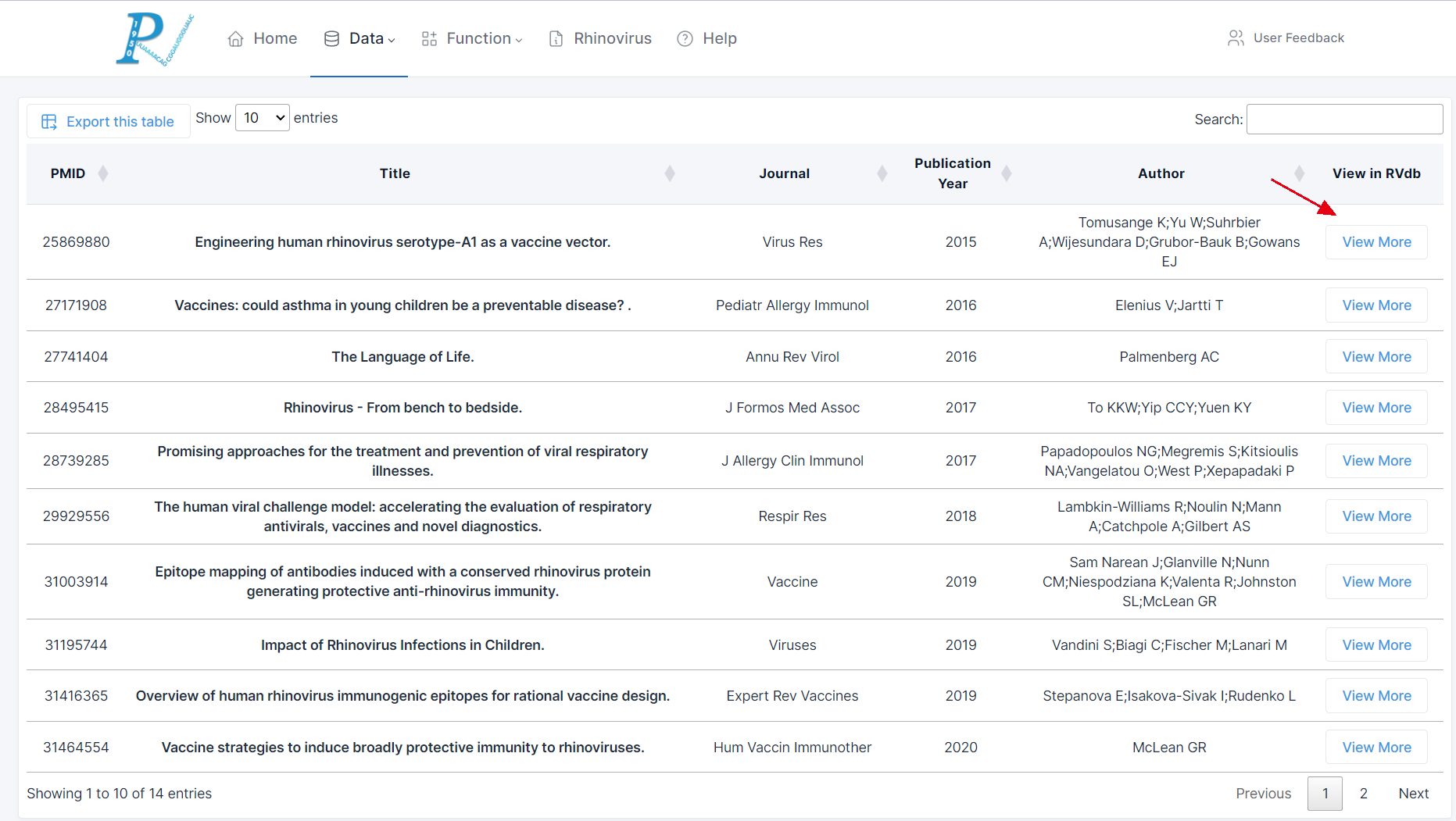
(5) The figure is an example of data retrieval: RV-related literature published since 2000 with the keyword "rhinovirus" and the MeSH term "vaccine".
(6) By clicking on "View More" in the last column, users can access the detailed publication information page.
(7) Within the publication information page, users have the ability to browse the publication content and click on specific "Keywords" or "MeSH terms" to discover other documents associated with the same keyword or MeSH term.

5.RVdb data retrieval error
If the following interface appears during data retrieval, there could be several possibilities:

(1) The data retrieval parameters are set incorrectly. Although we set up a comprehensive submission verification function as much as possible before submitting the form to ensure the correctness of the data submitted by the user, it is still possible that the retrieval parameters submitted by the user are incorrect.
(2) Target data record not found: It is possible that the specific data record being searched for is not available in RVdb.
(3) RVdb updating and optimizing: The interface could appear during RVdb's internal data updates and optimizations.
To resolve the issue, you can try re-querying the data by ensuring the query parameters are accurate or optimizing your query strategy. If the problem persists, it is recommended to contact the RVdb development team at zhaopeng0539@126.com for further assistance.
Online analysis function
1.How to perform RVdb online multiple sequence alignment?
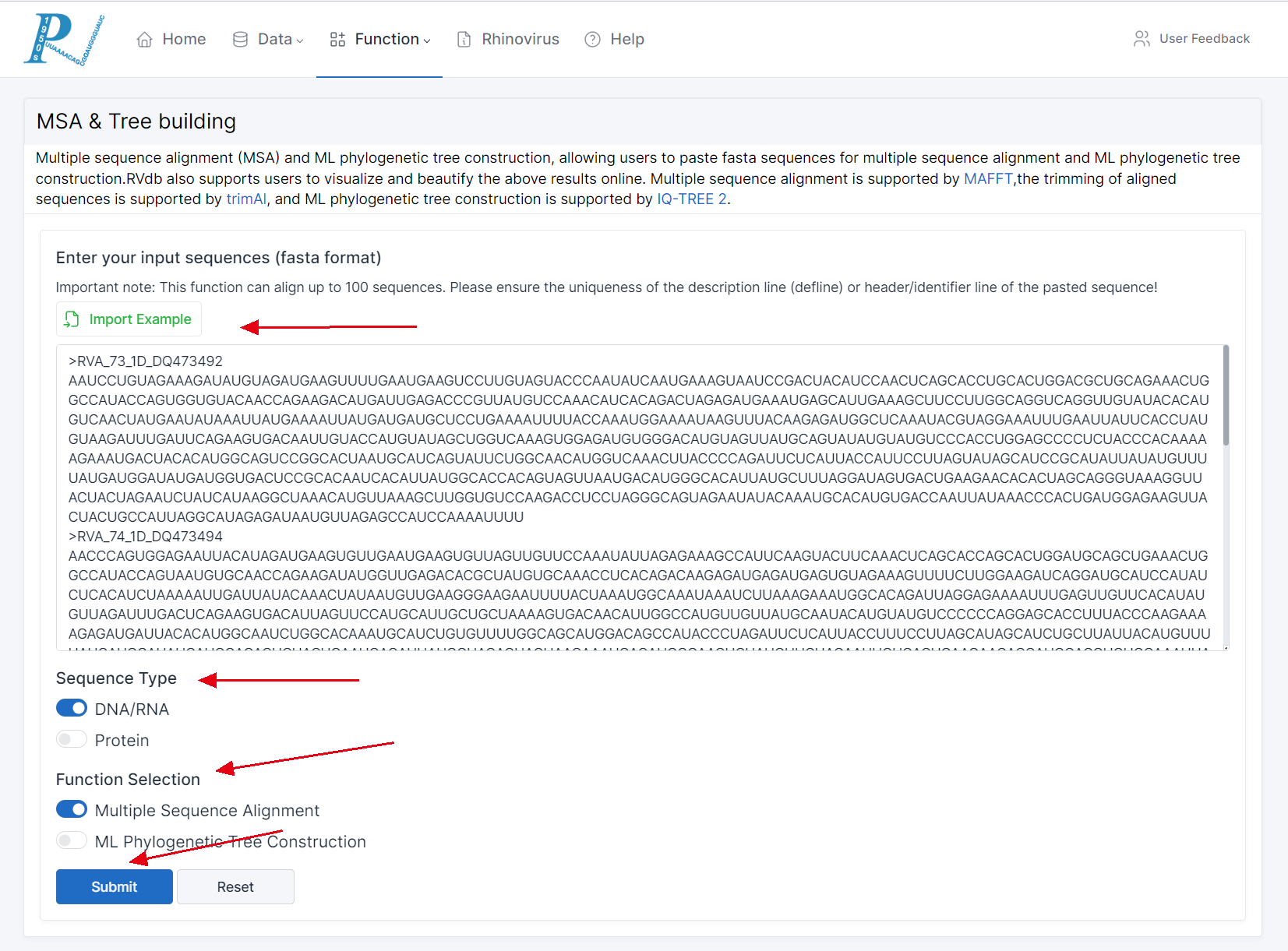
(1) Multiple sequence alignment (MSA) and ML phylogenetic tree construction, allowing users to paste fasta sequences for multiple sequence alignment and ML phylogenetic tree construction.RVdb also supports users to visualize the above results online. Multiple sequence alignment is supported by MAFFT,the trimming of aligned sequences is supported by trimAl, and ML phylogenetic tree construction is supported by IQ-TREE 2.
(2) Enter the sequences to be aligned in fasta format in the input box.
(3) Ensure the number of sequences is between 4 and 100.
(4) Before inputting, make sure that the headers of these fasta format sequences (the text after ">") are unique. Although we have set up a verification function before form submission to alert users, sequences with duplicate headers can cause analytical errors if passed to the RVdb backend.
(5) Select "Sequence Type". If the selected sequence type does not match the uploaded sequence, RVdb will also throw an error.
(6) Finally, in the "Function Selection" section, choose "Multiple Sequence Alignment" and click "Submit".
(7) If you wish to try this function, please click "Import Example" and complete the above steps.
(8) RVdb will return a multi-sequence alignment visualization page based on MSAViewer to the user. Users can carry out visualization, result exportation, and other operations on the alignment results.

2.How to build a phylogenetic tree fully automatically on RVdb?
(1) Multiple sequence alignment (MSA) and ML phylogenetic tree construction, allowing users to paste fasta sequences for multiple sequence alignment and ML phylogenetic tree construction.RVdb also supports users to visualize the above results online. Multiple sequence alignment is supported by MAFFT,the trimming of aligned sequences is supported by trimAl, and ML phylogenetic tree construction is supported by IQ-TREE 2.
(2) Enter the sequences to be aligned, in fasta format, into the input box.
(3) Ensure that the number of sequences falls within the range of 4 to 100.
(4) Before inputting, please make sure that the headers of these fasta format sequences (the text after the ">") are unique. While we have set a validation function before form submission to notify users, sequences with duplicate headers could lead to analytical errors when passed to the RVdb backend.
(5) Select "Sequence Type". Please note that if the selected sequence type does not match the uploaded sequences, RVdb will return an error.
(6) Finally, in the "Function Selection", choose "ML Phylogenetic Tree Construction" and click "Submit".
(7) If you wish to try out these features, please click on "Import Example" and complete the above steps.
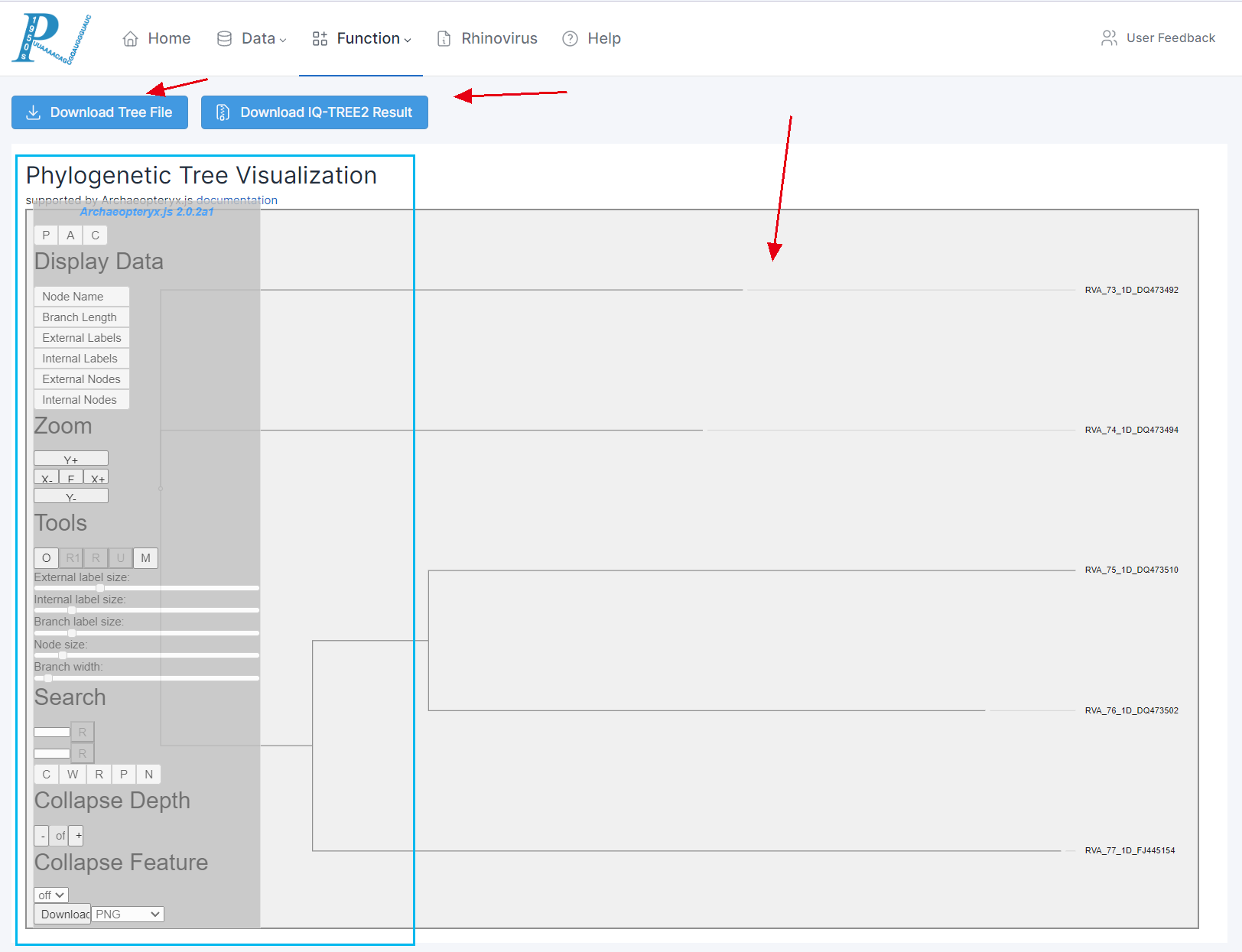
(8) RVdb will provide the user with an interactive online system for visualizing phylogenetic trees. Users can opt to download the tree files, download all the result files from IQtree2, or carry out a series of visualization and file export operations through Archaeopteryx.js. For more information, please visit the help documentation
3.How to perform RVdb BLAST analysis?

(1) First, users should choose an appropriate blast program based on their analysis needs. This tutorial uses "blastn" as an example.
(2) Paste a fasta-formatted sequence to be analyzed.
(3) Set the appropriate parameters. The meaning of the parameters can be referenced here.
(4) If you're unsure about the specific parameters you're using, it's recommended to stick with the default parameters provided by RVdb BLAST.
(5) Afterwards, click "submit".
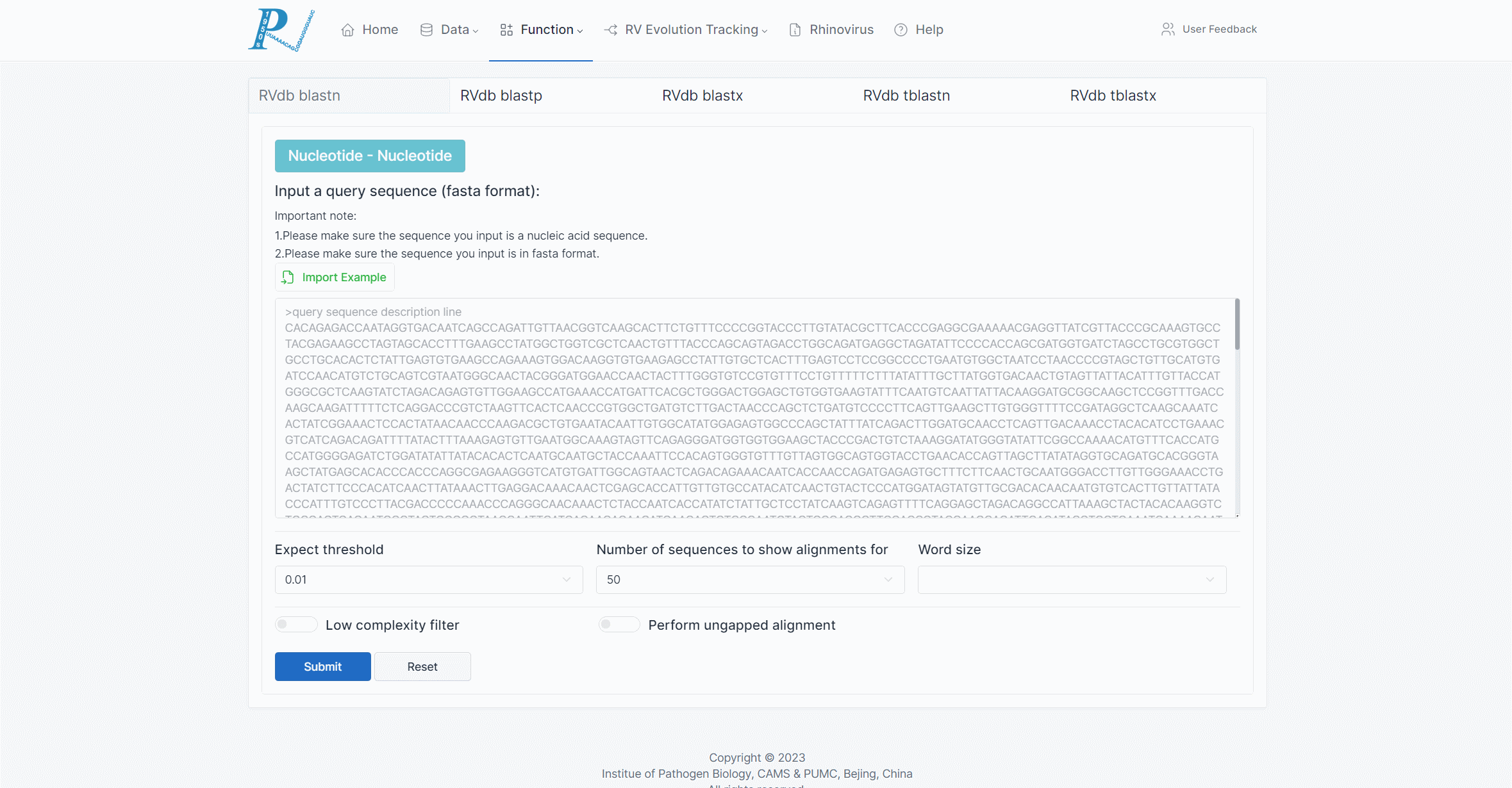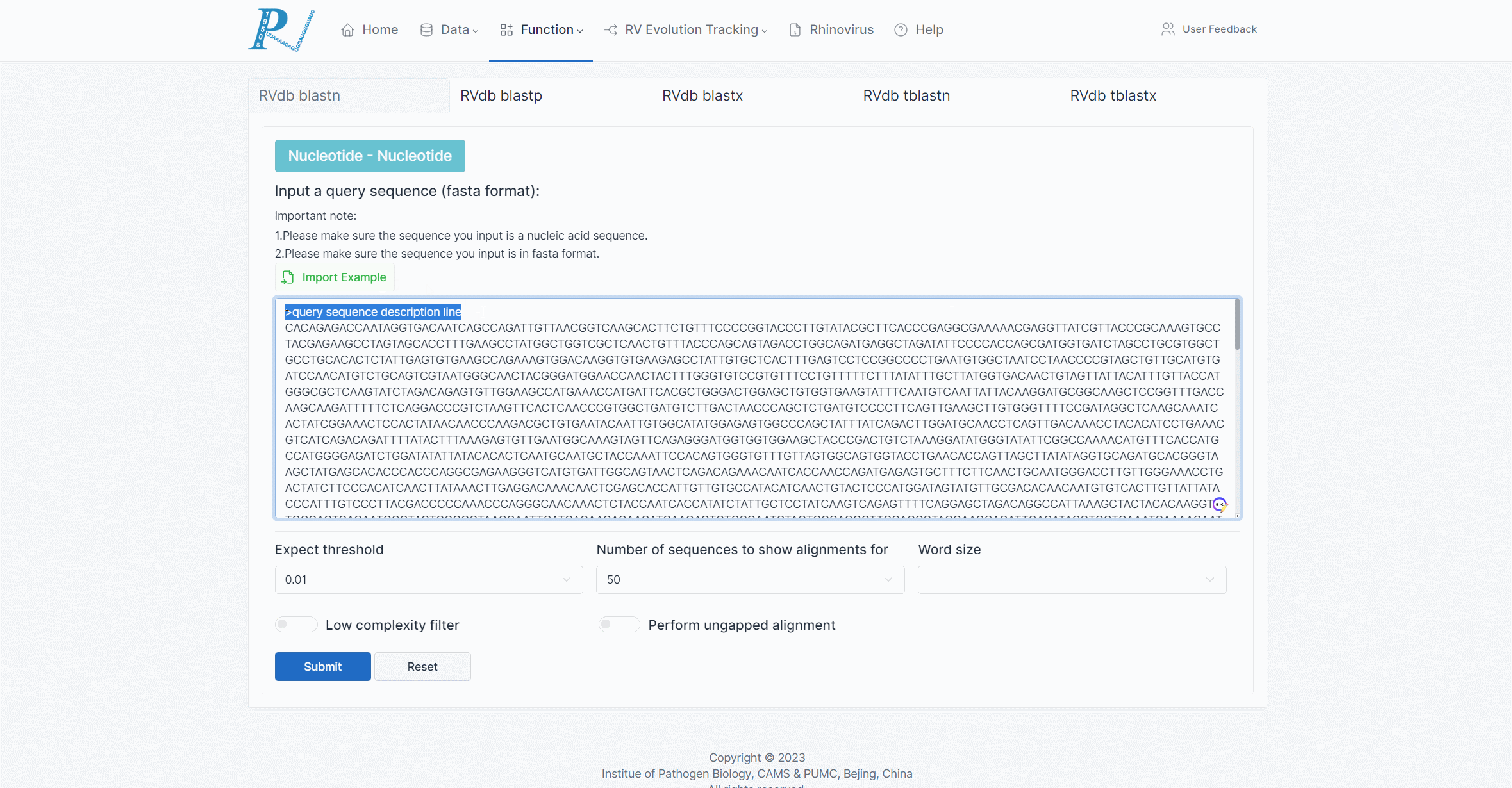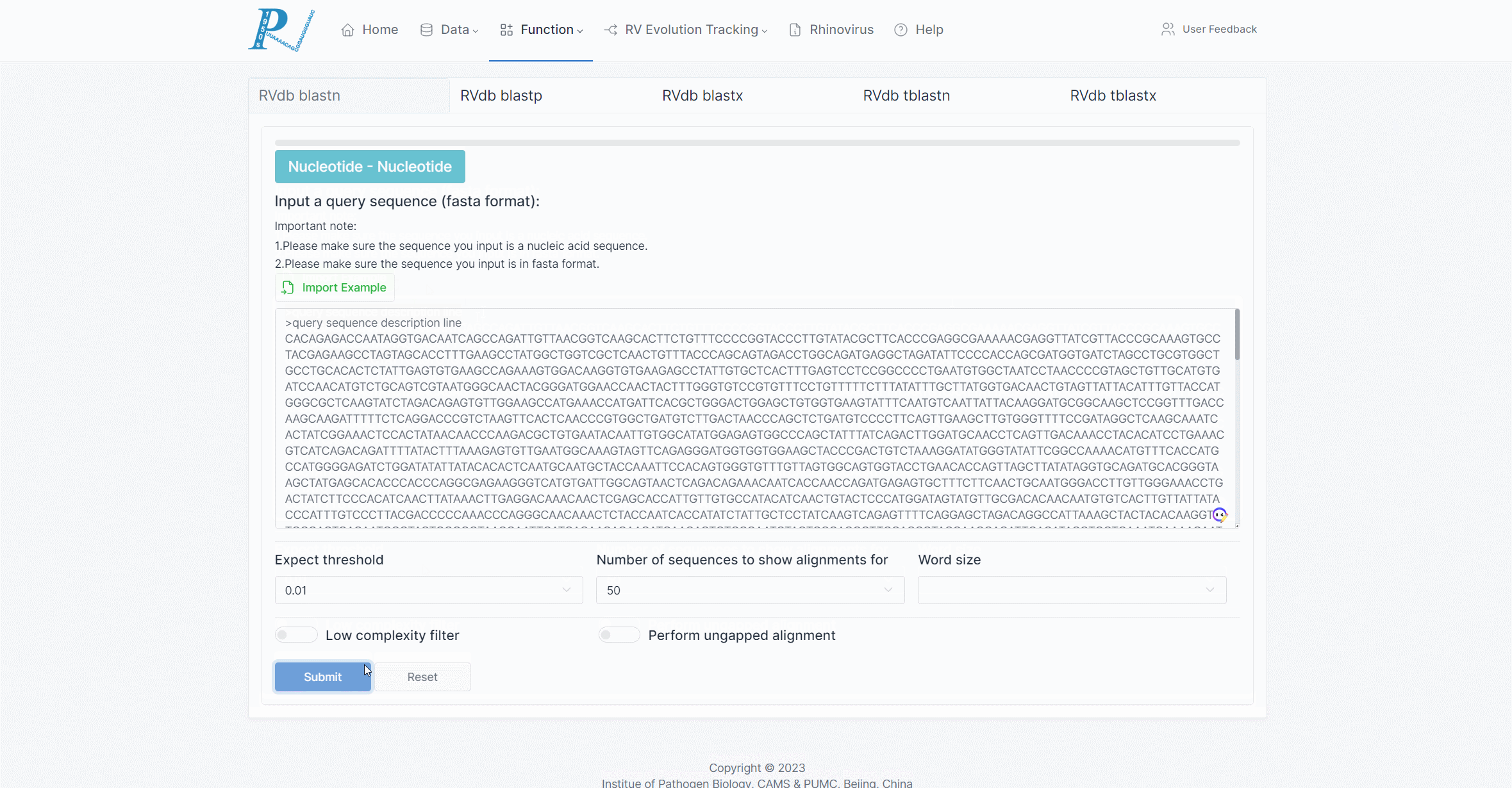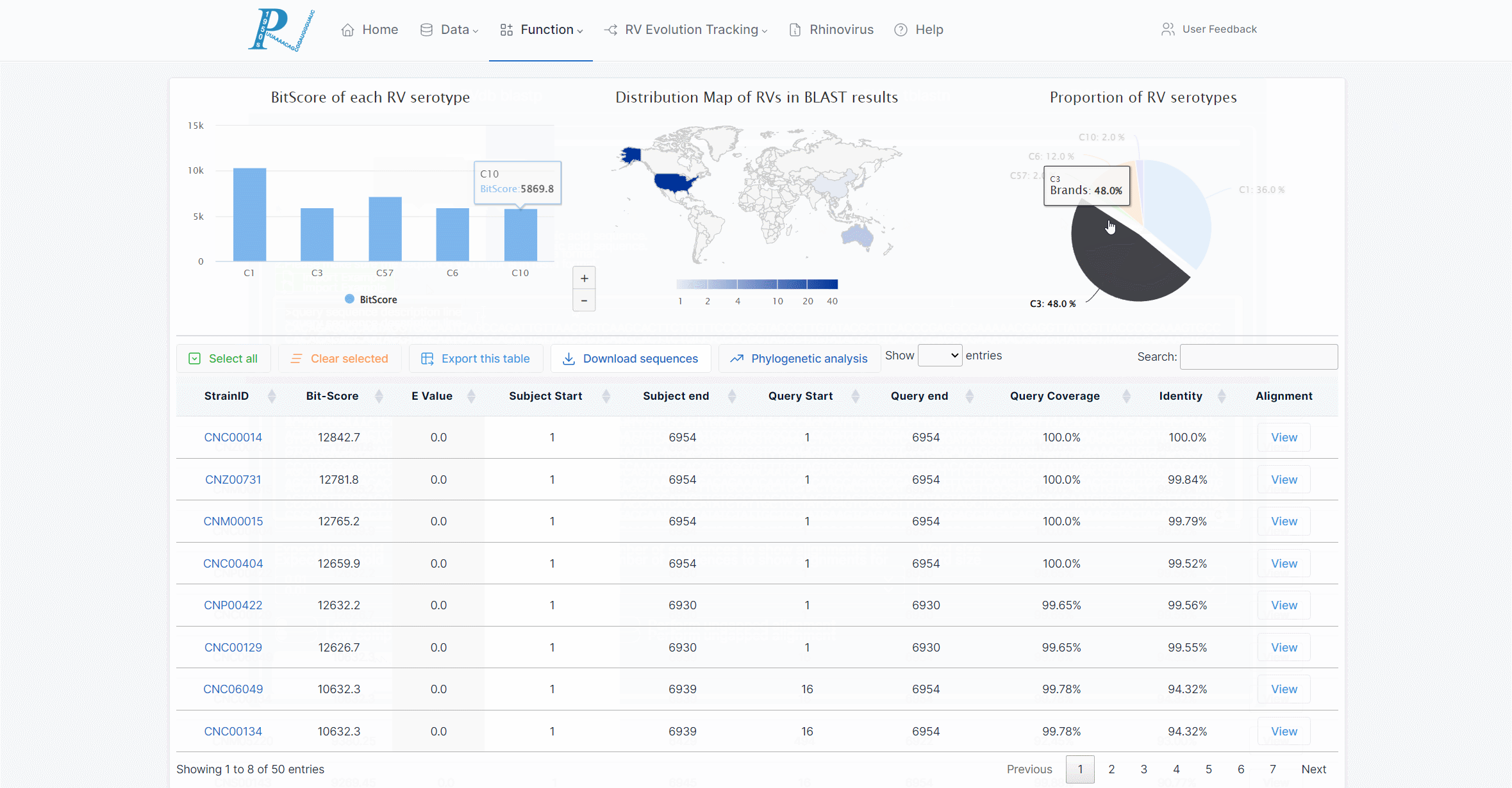
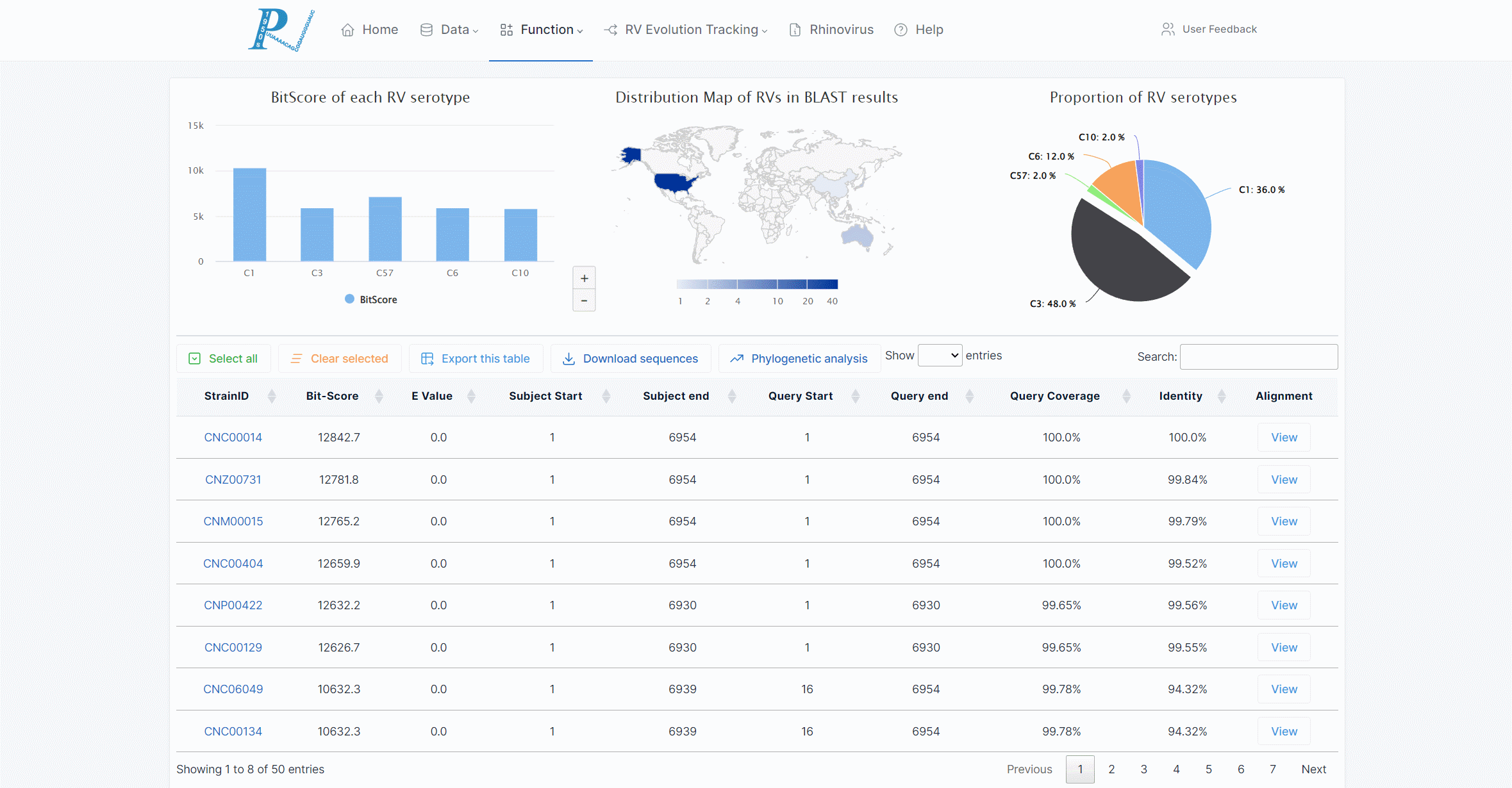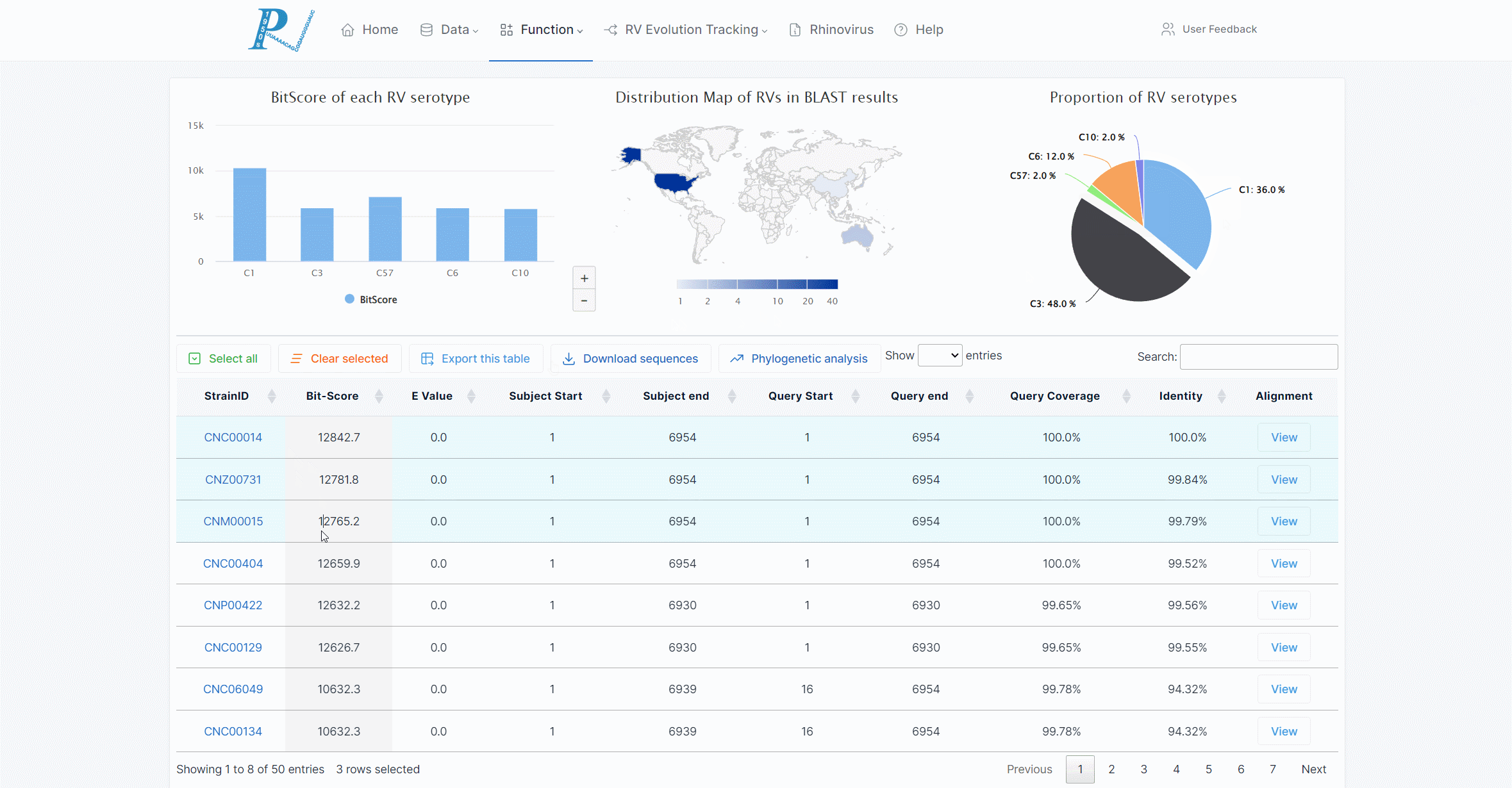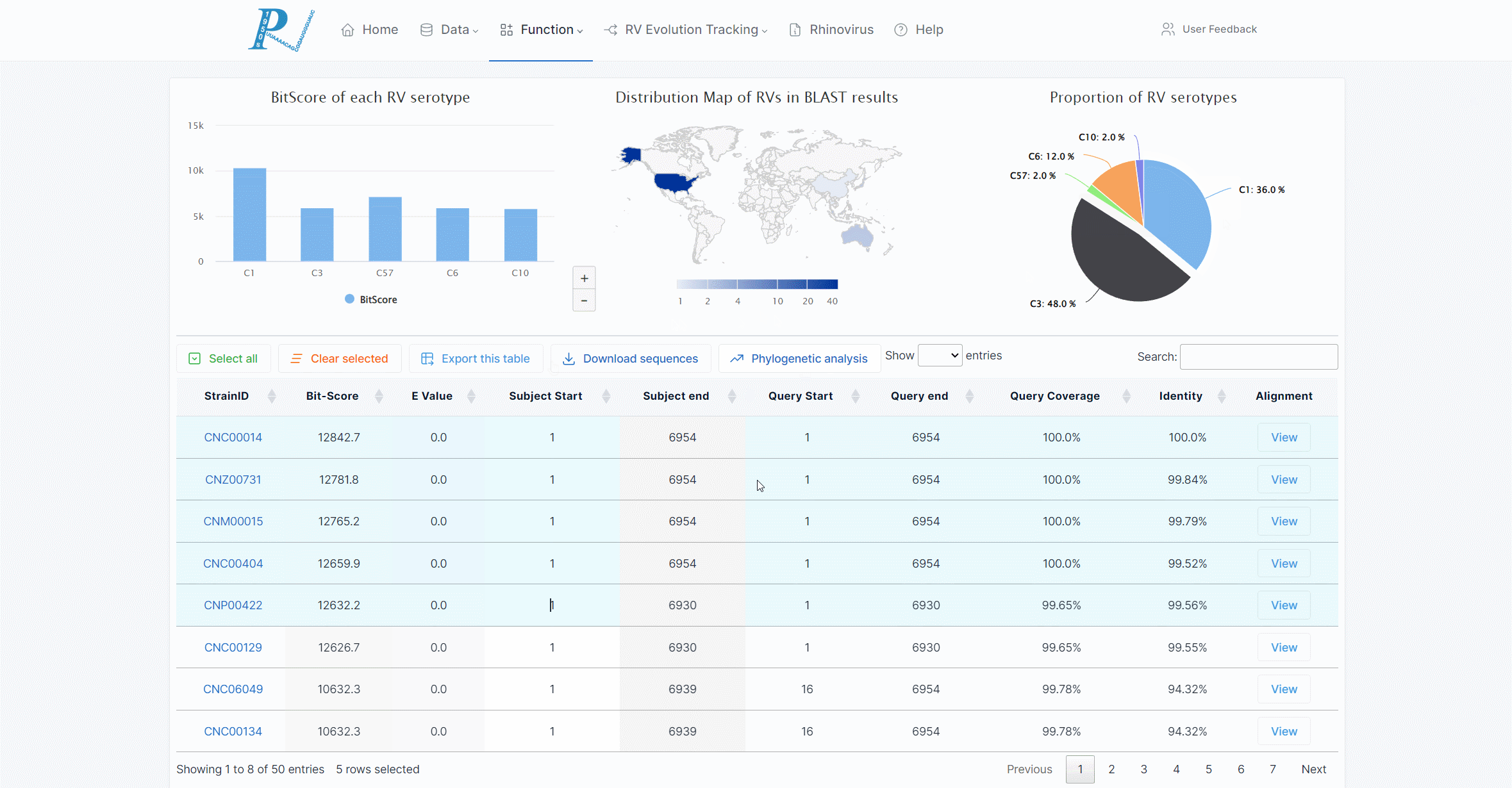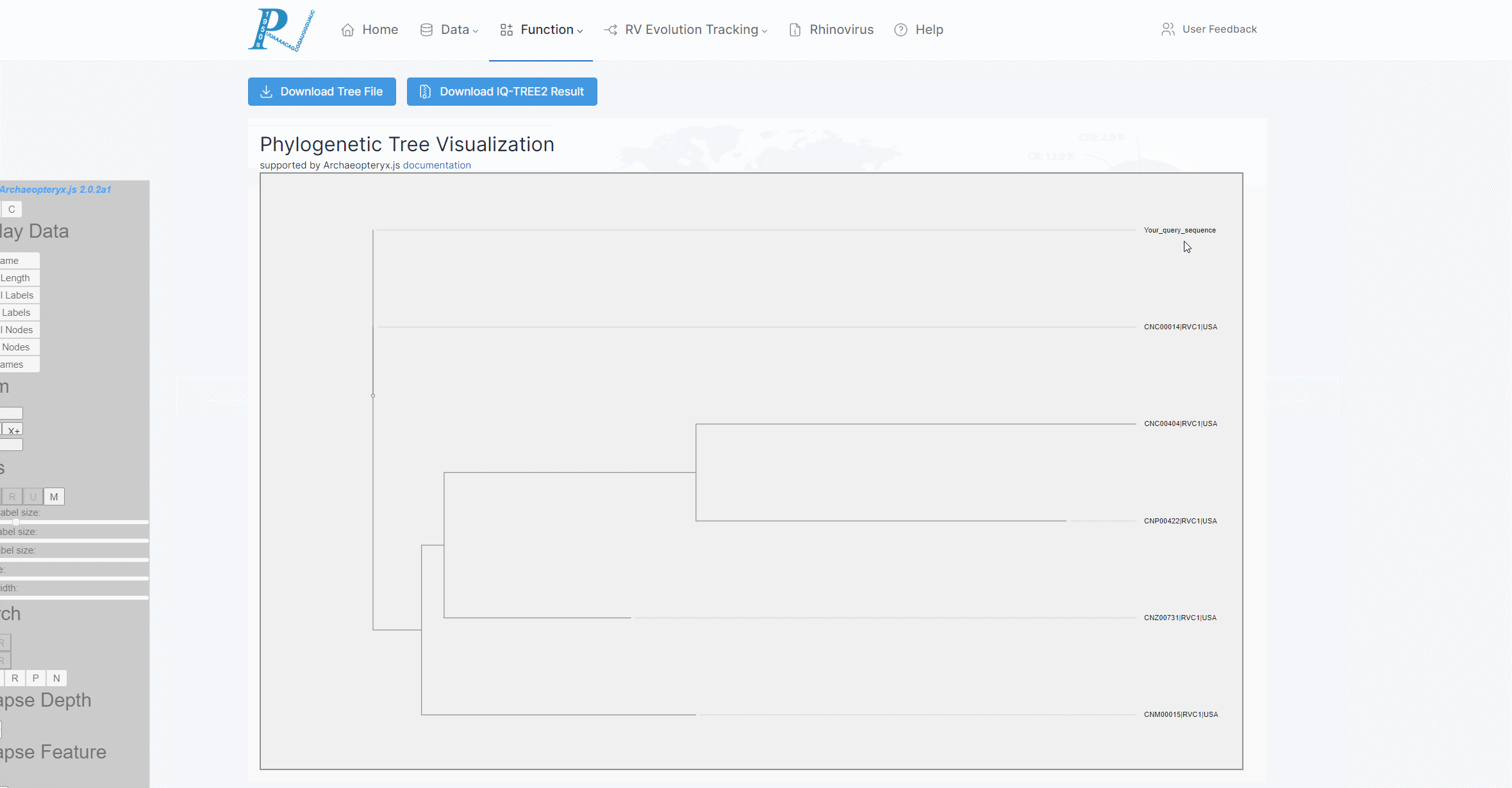
(6) The returned interactive graphics include the average BLAST score (BitScore) in each serotype, the global distribution of RV strains in the results, and the proportion of serotypes.
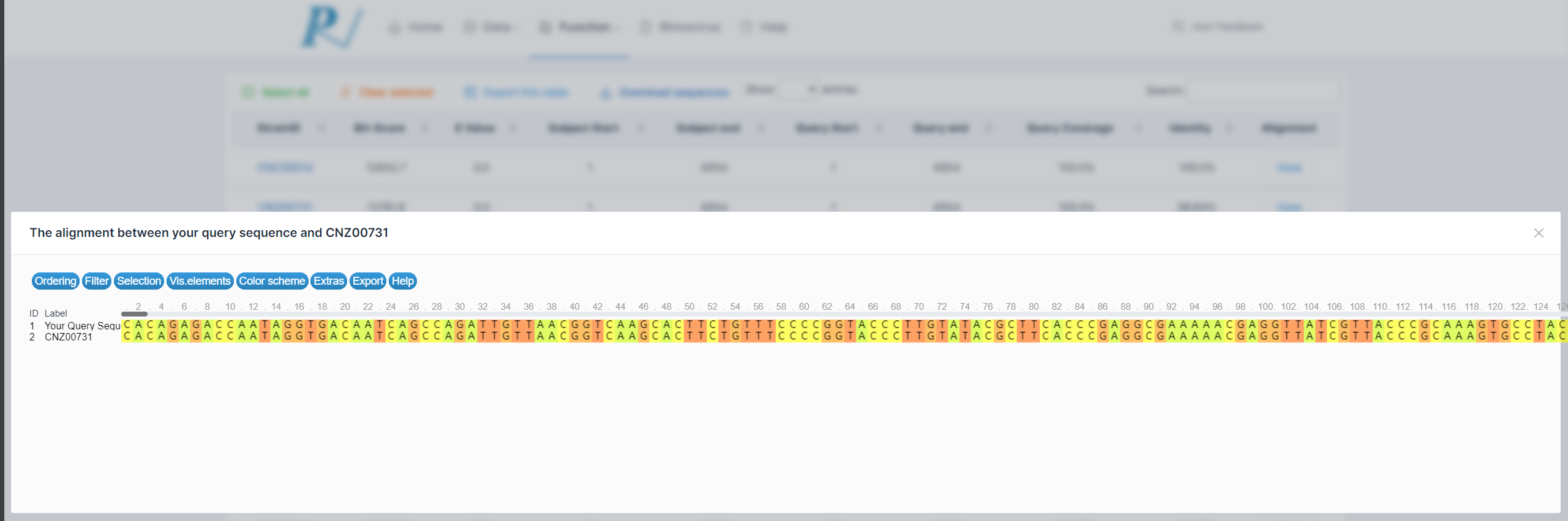
(7) The returned results will document the alignment with the viral strains in the database. Users can export the table or view the alignment results online.
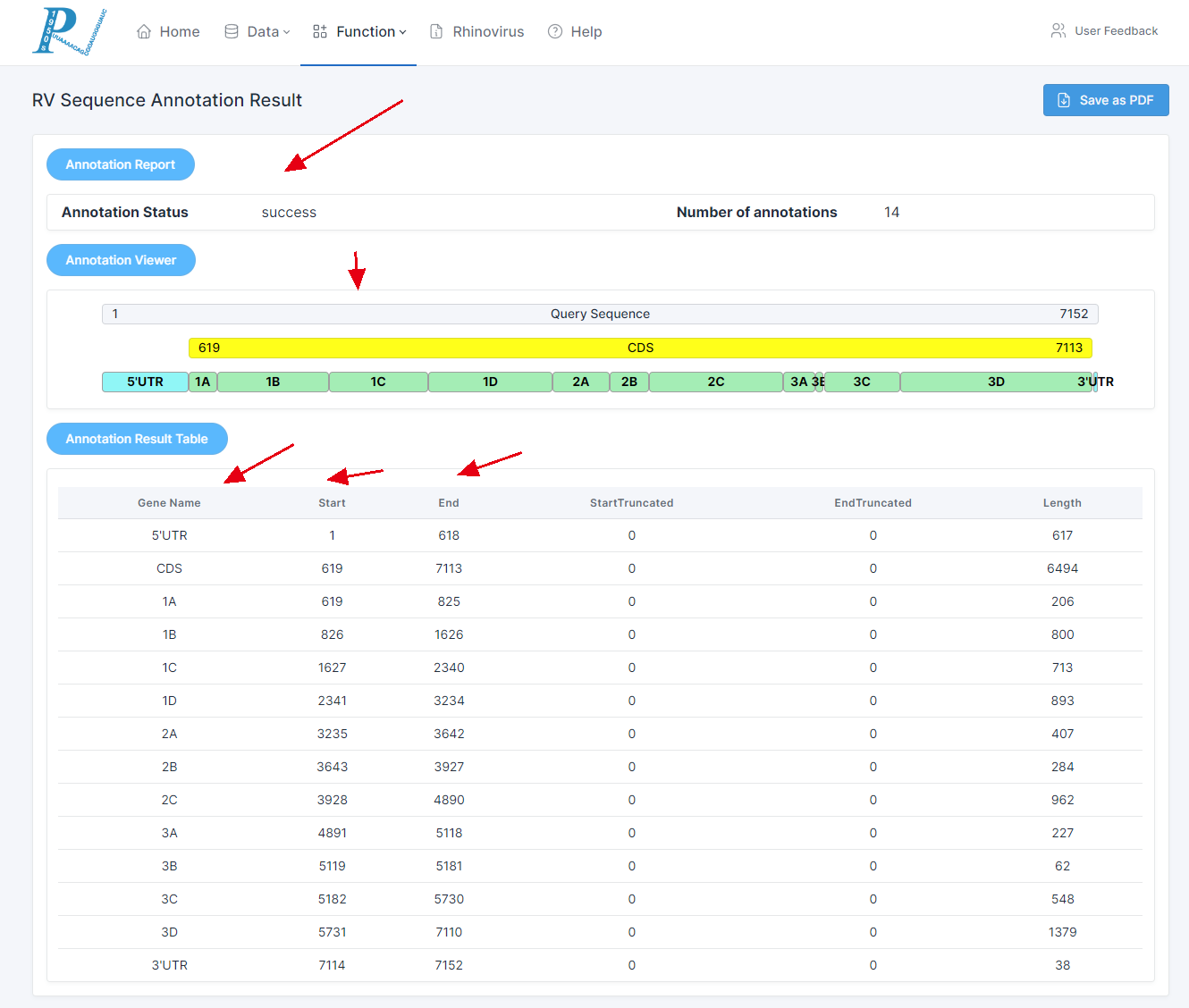
4.How to perform RV sequence annotation analysis?
(1) The operation process of RV sequence annotation is very straightforward. Users only need to input one rhinovirus sequence in fasta format that is to be annotated. If the sequence isn't a rhinovirus sequence, the annotation might be unsuccessful due to low similarity.
(2) Click "submit" to query the annotation results.
(3) The report interface includes, in order, the annotation report, sequence annotation visualization, and annotation information table.
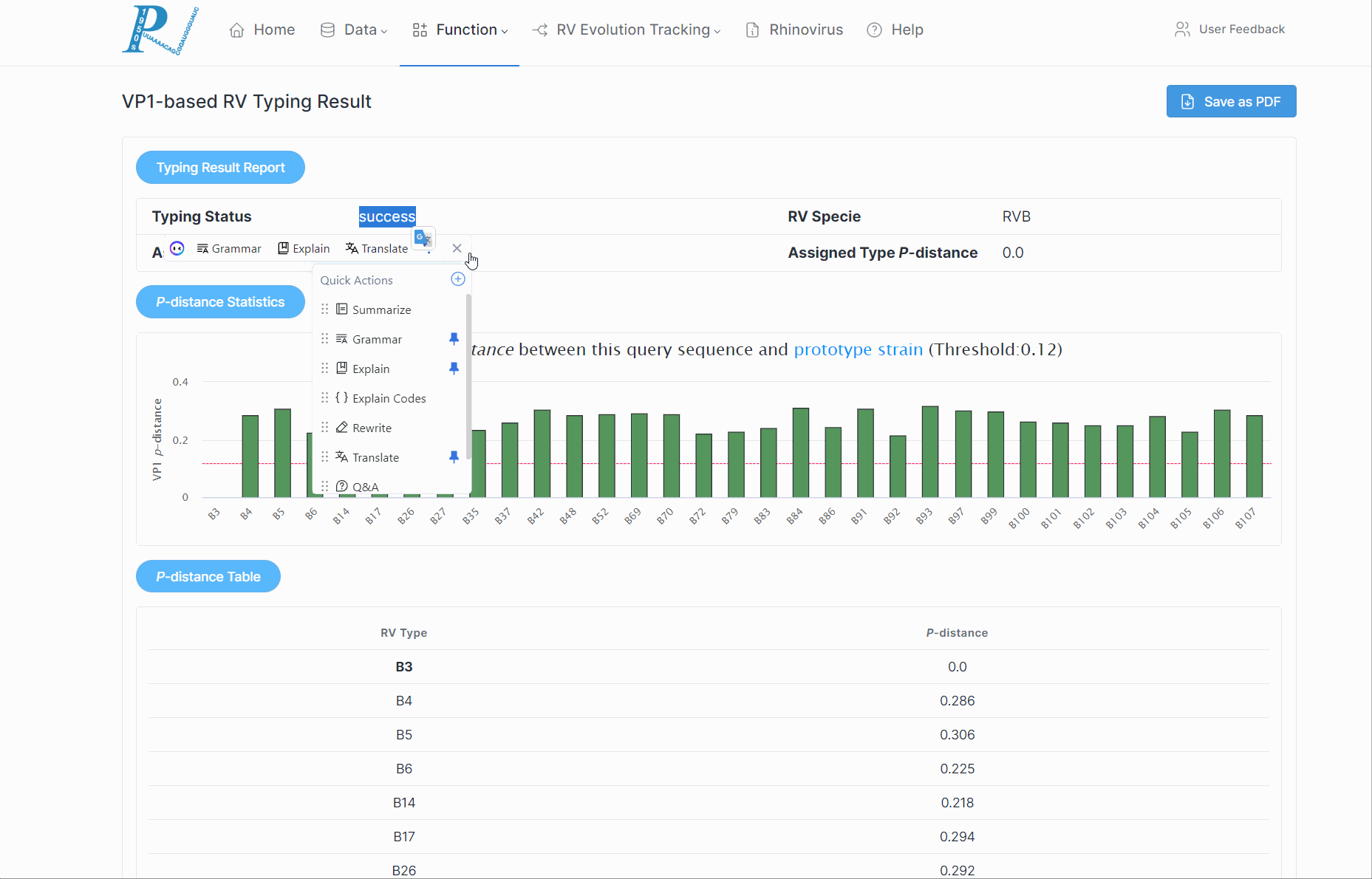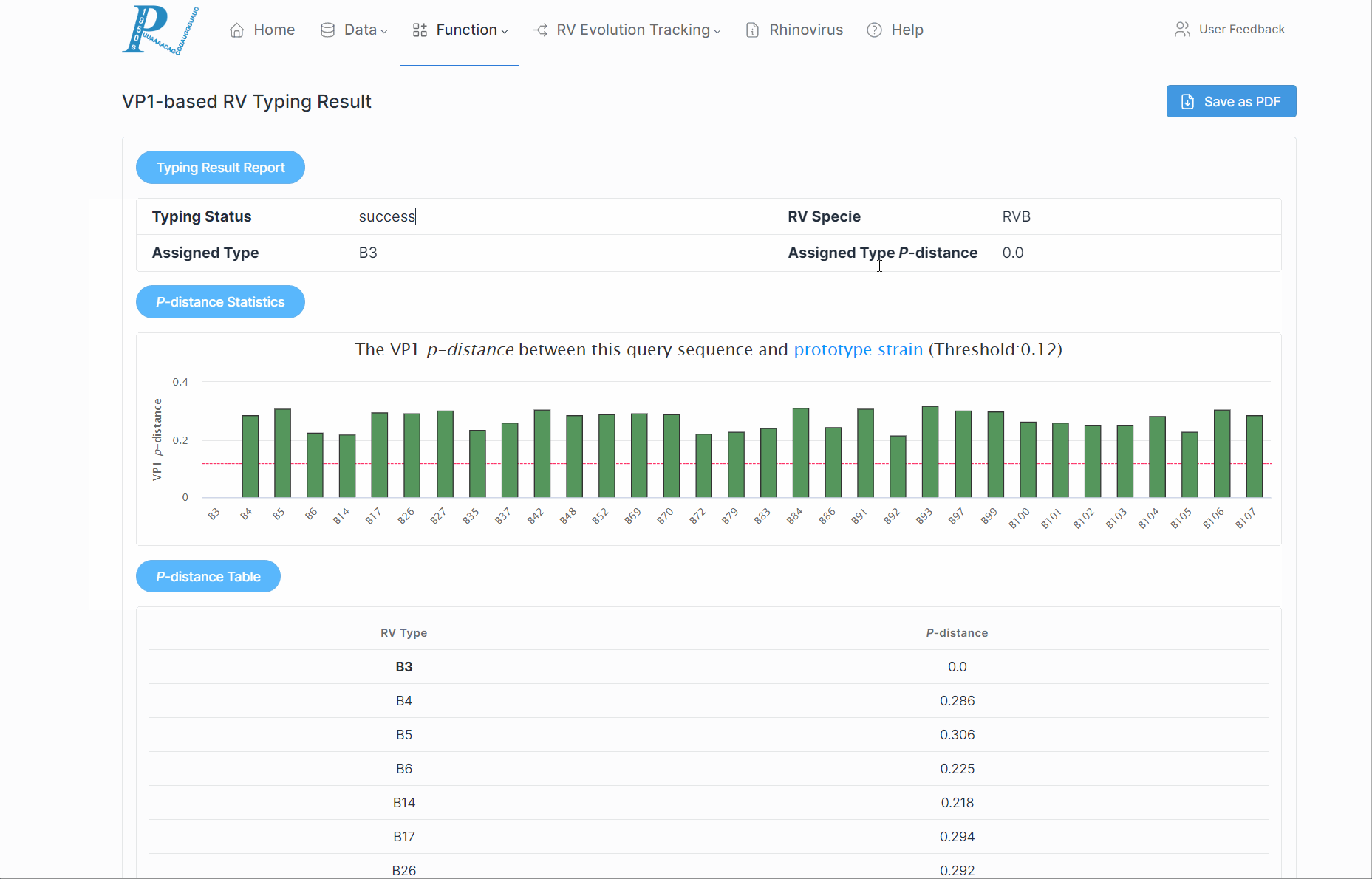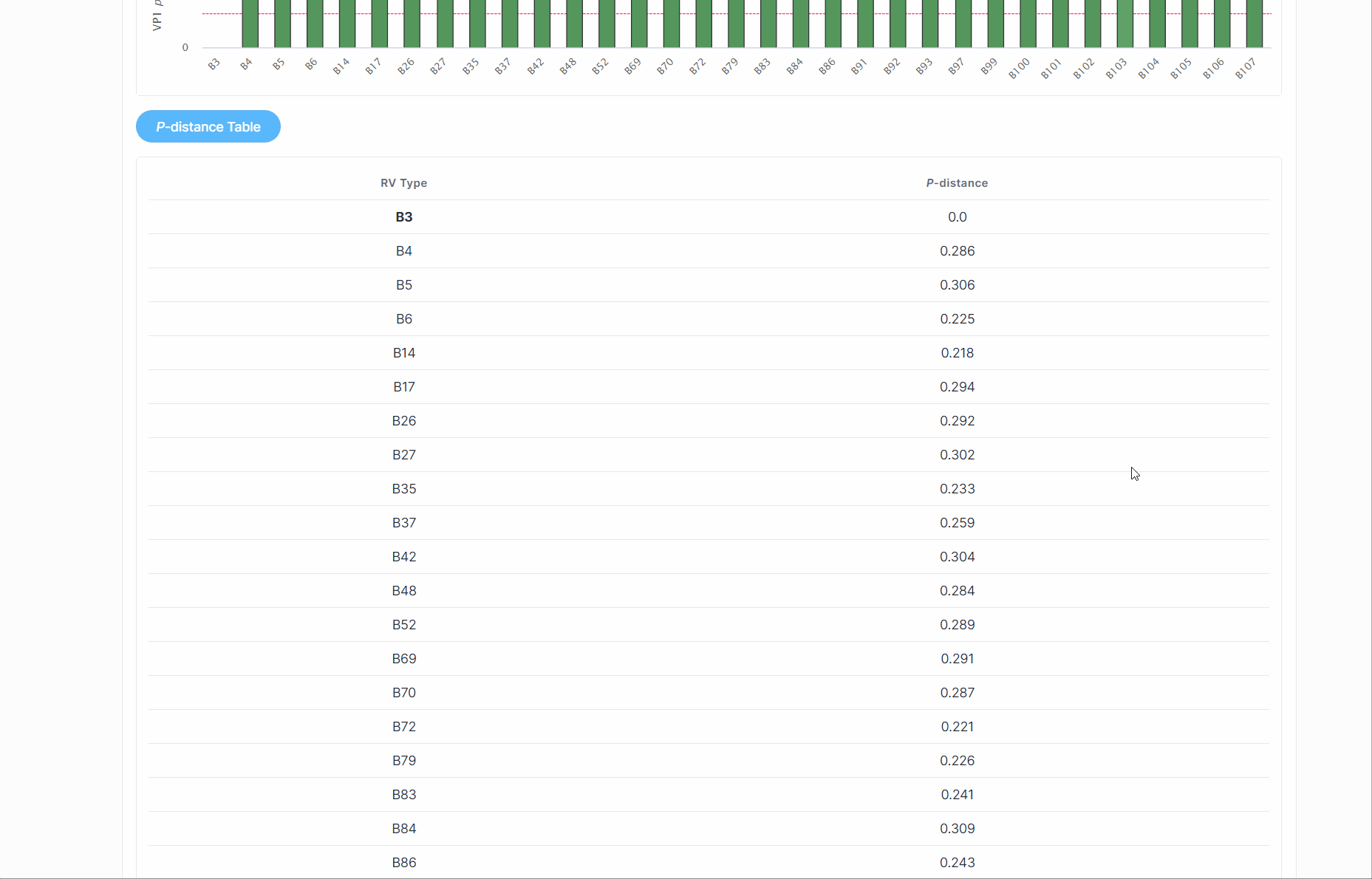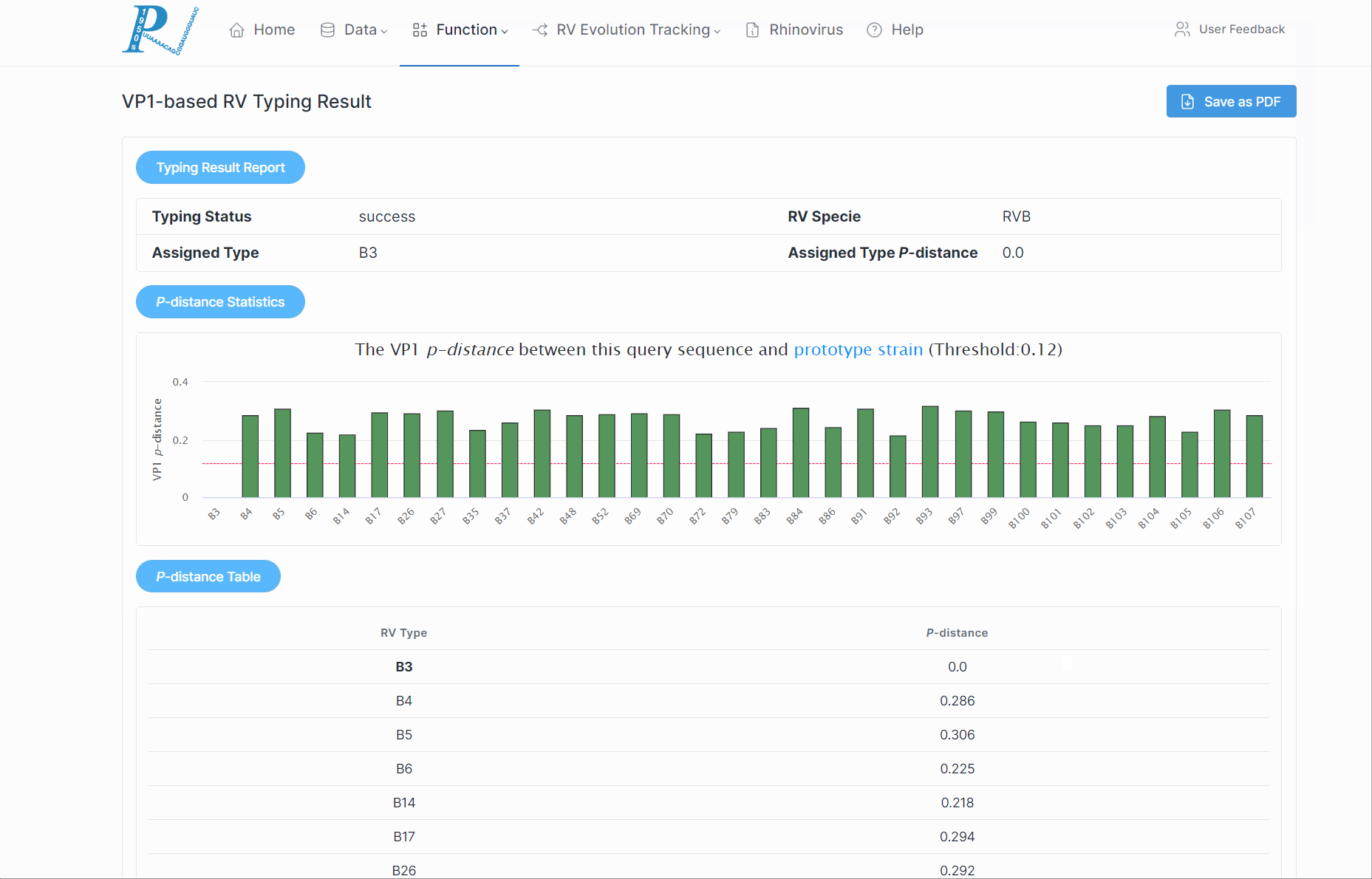
5.How to perform rhinovirus molecular typing analysis?
(1) First, ensure the input is a VP1/VP4/2 nucleotide sequence and is in fasta format.
(2) Click "Submit". This process may take some time, so please be patient.
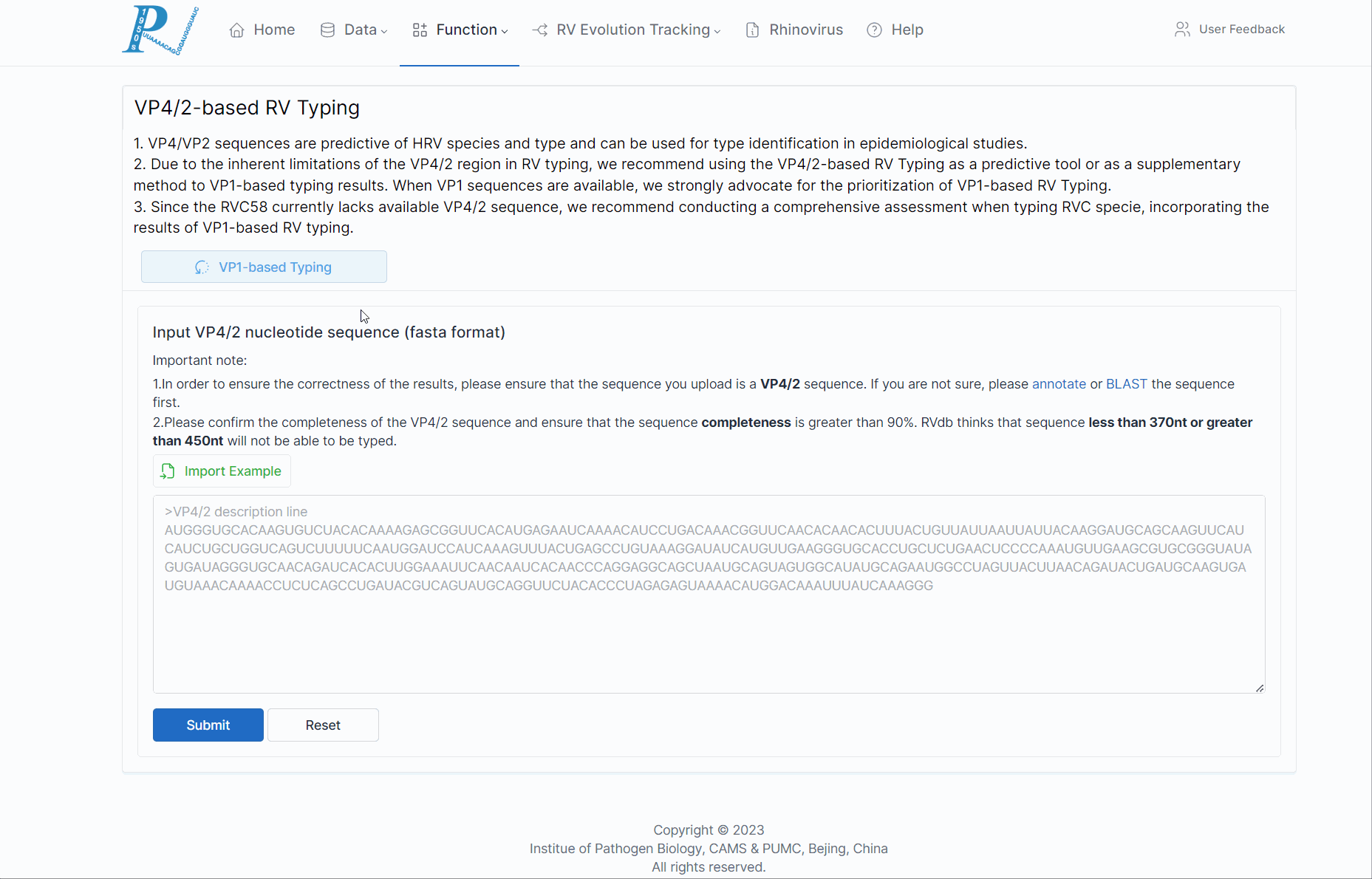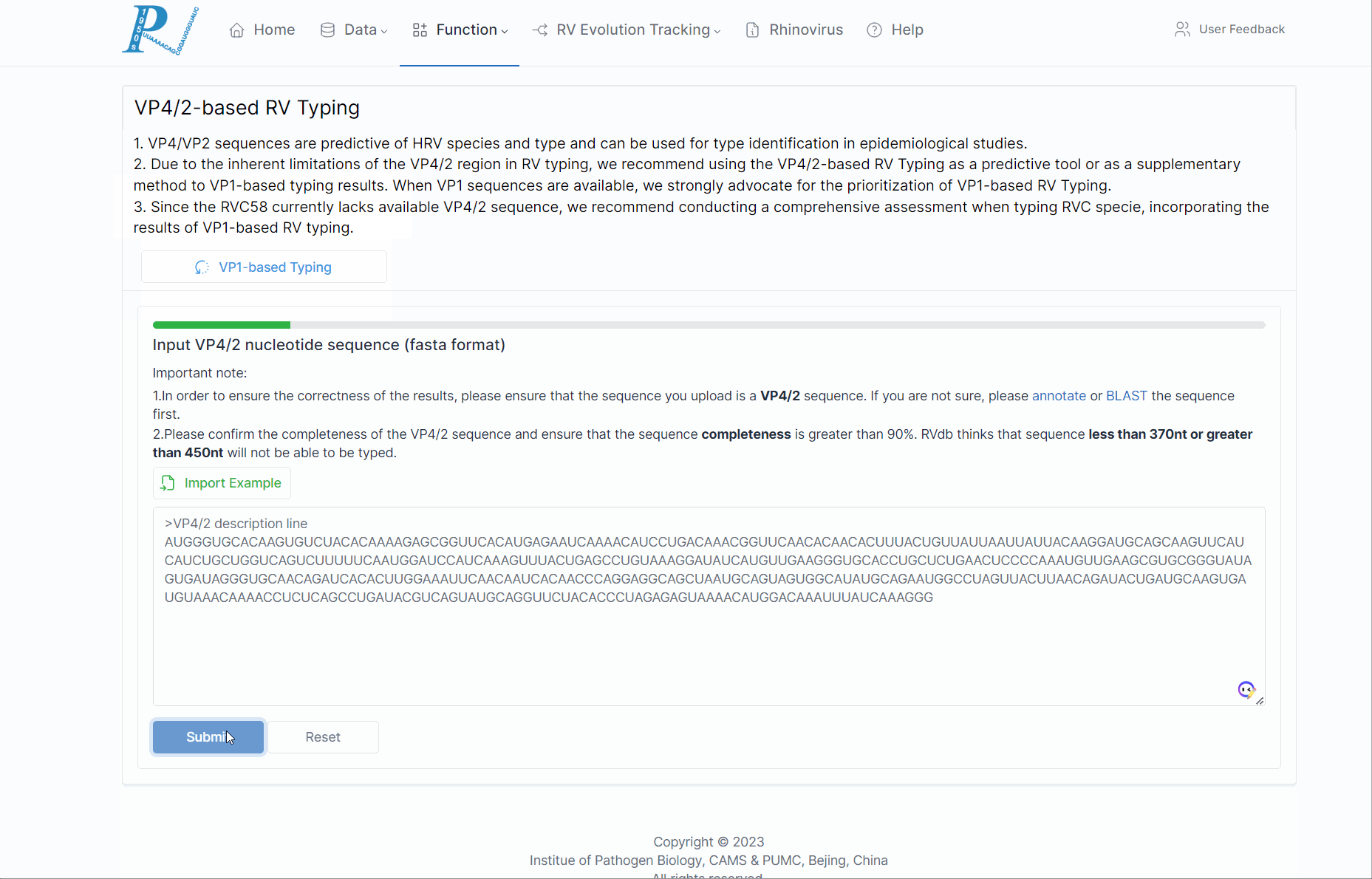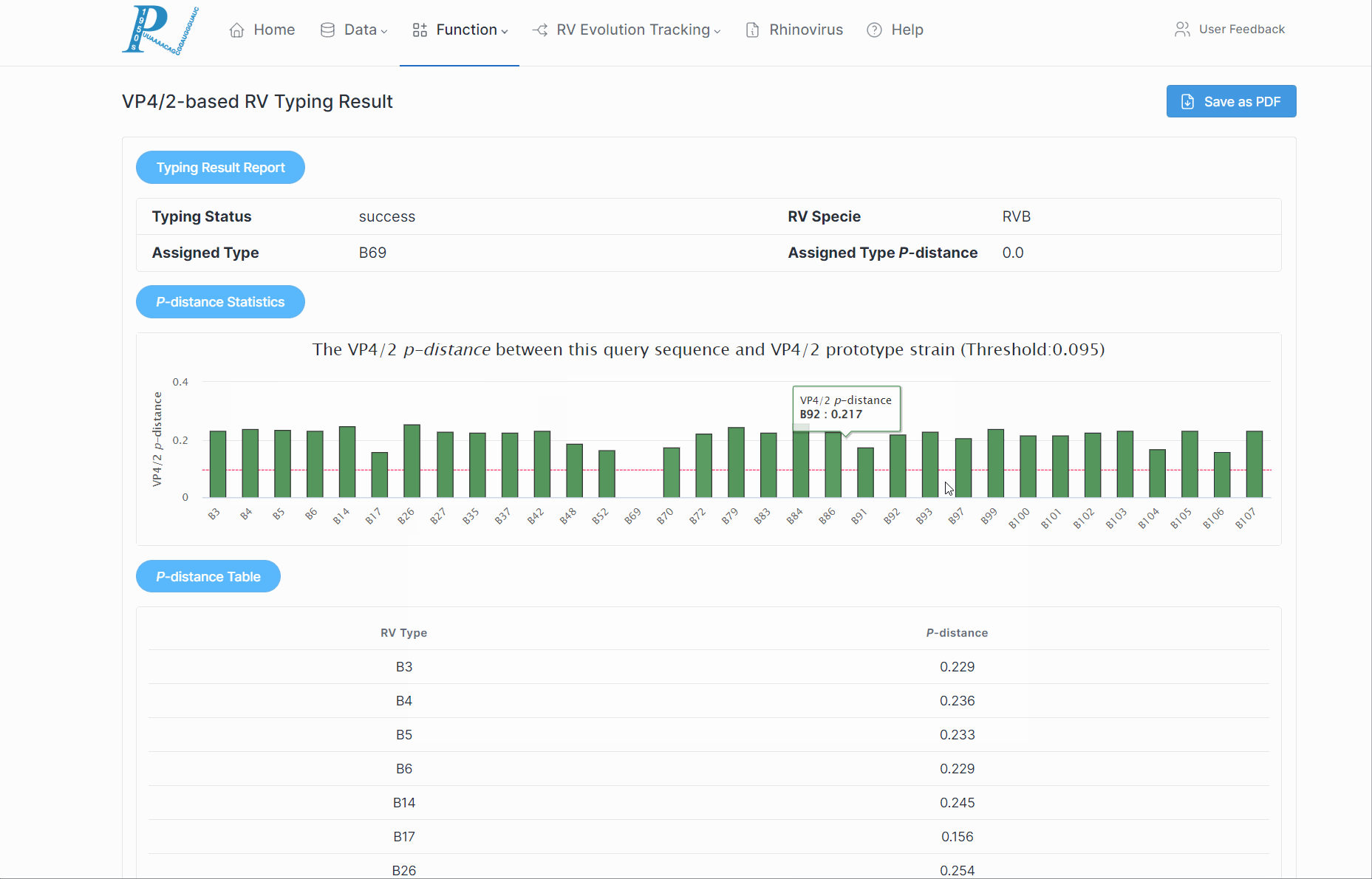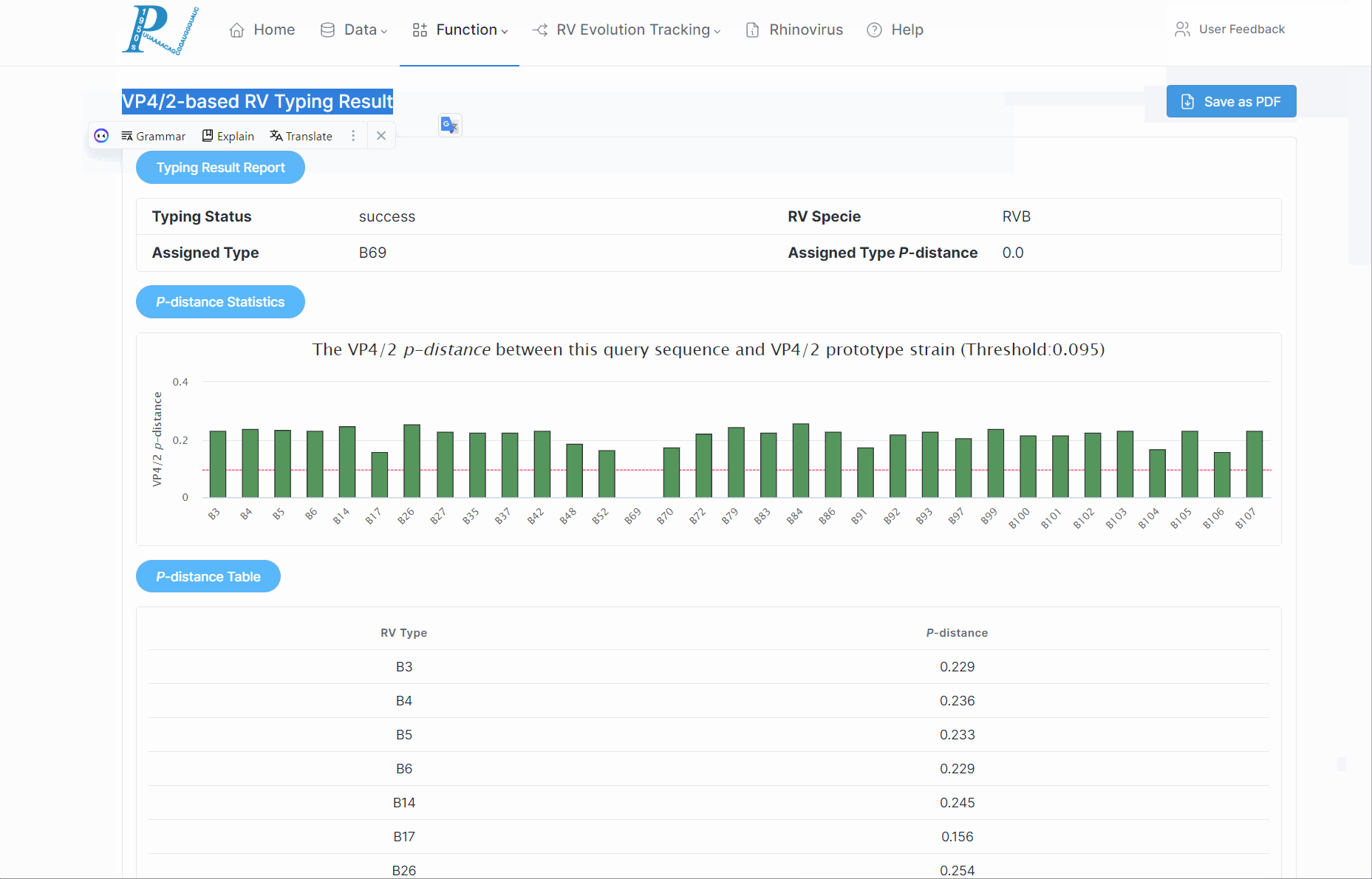
(3) Subsequently, RVdb will return a serotyping report to the user, along with the p-distance of various serotypes within the RV species.
(4) If a potential new serotype is discovered, users should further combine to the phylogenetic relationships.